Cancerous melanoma cells, shown with their cell bodies (green) and nuclei (blue), are nestled in tiny hollow lumens (tubes) within the cryogel (red) structure. (credits: Thomas Ferrante, Sidi A. Bencherif / Wyss Institute at Harvard University)
A new biologically inspired “injectable cryogel whole-cell cancer vaccine” combines patient-specific harvested cancer cells and immune-stimulating chemicals or biological molecules to help the body attack cancer. It has been developed by scientists at Harvard’s Wyss Institute and Dana-Farber Cancer Institute.
This new approach is simpler and more economical than other cancer cell transplantation therapies, which harvest tumor cells and then genetically engineer them to trigger immune responses once they are transplanted back into the patient’s body, the researchers say.
The research, headed by Wyss Core Faculty member David Mooney, Ph.D., was reported online in an open-access paper in Nature Communications on August 12.
Minimally invasive cryogels
The new anti-cancer vaccine uses the patient’s own cancer cells to trigger immune responses. The cryogels are a type of hydrogel made up of hydrophilic (water-compatible) polymer chains that are cross-linked and can hold up to 99 percent water.
They are created by freezing a solution of the polymer that is in the process of gelling. When thawed back again to room temperature, the substance turns into a highly interconnected pore-containing hydrogel, which is similar in composition to bodily soft tissues in terms of their water content, structure, and mechanics.

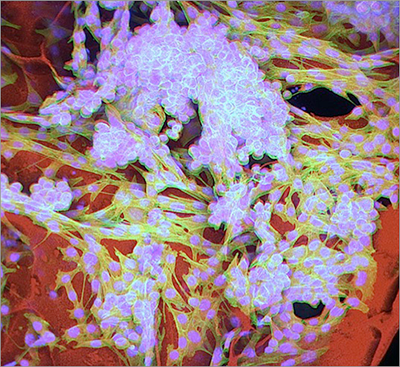
This scanning electron microscopy image shows the thawed cryogel with its well-organized interconnected porous architecture ready to be infused with cancer cells and immune factors. (credits: Ellen Roche, James Weaver, Sidi A. Bencherif / Wyss Institute at Harvard University)
By adjusting their shape, physical properties, and chemical composition, Mooney’s team generated sponge-like, porous cryogels that can be infused with living cells, biological molecules, or drugs for a variety of potential therapeutic applications, including cancer immunotherapy.
The cryogels are minimally invasive because of their extreme flexibility and resilience, enabling them to be compressed to a fraction of their size and injected underneath the skin via a surgical needle. Once injected, they quickly bounce back to their original dimensions to do their job.
“After injection into the body, the cryogels can release their immune-enhancing factors in a highly controlled fashion to recruit specialized immune cells, which then make contact and read unique signatures off the patient’s tumor cells, also contained in the cryogels,” said Sidi Bencherif, the study’s co-first author and a Research Associate in Mooney’s research group.
This has two consequences, he said: “Immune cells become primed to mount a robust and destructive response against patient-specific tumor tissue and the immune tolerance developing within the tumor microenvironment is broken.”
Shrinking tumors
In experimental animal models on melanoma (skin) tumors, results show that using the cryogel to deliver whole cells and drugs triggers a dramatic immune response that can shrink tumors and even prophylactically (in advance) protect animals from tumor growth. With the pre-clinical success of the new cancer cell vaccination technology, Mooney and his team are going to explore how this cryogel-based method could be more broadly useful to treat a number of different cancer types.
“This new injectable form of this biomaterials-based cancer vaccine will help to expand the cancer immunotherapy arsenal, and it’s a great example of how engineering and materials science can be used to mimic the body’s own natural responses in a truly powerful way,” said Don Ingber, the Wyss Institute’s Founding Director, who also is the Judah Folkman Professor of Vascular Biology at Harvard Medical School and Boston Children’s Hospital, and Professor of Bioengineering at SEAS.
Mooney is also the Robert P. Pinkas Family Professor of Bioengineering at the Harvard John A. Paulson School of Engineering and Applied Sciences.
Abstract of Automated adaptive inference of phenomenological dynamical models
Dynamics of complex systems is often driven by large and intricate networks of microscopic interactions, whose sheer size obfuscates understanding. With limited experimental data, many parameters of such dynamics are unknown, and thus detailed, mechanistic models risk overfitting and making faulty predictions. At the other extreme, simple ad hoc models often miss defining features of the underlying systems. Here we develop an approach that instead constructs phenomenological, coarse-grained models of network dynamics that automatically adapt their complexity to the available data. Such adaptive models produce accurate predictions even when microscopic details are unknown. The approach is computationally tractable, even for a relatively large number of dynamical variables. Using simulated data, it correctly infers the phase space structure for planetary motion, avoids overfitting in a biological signalling system and produces accurate predictions for yeast glycolysis with tens of data points and over half of the interacting species unobserved.
