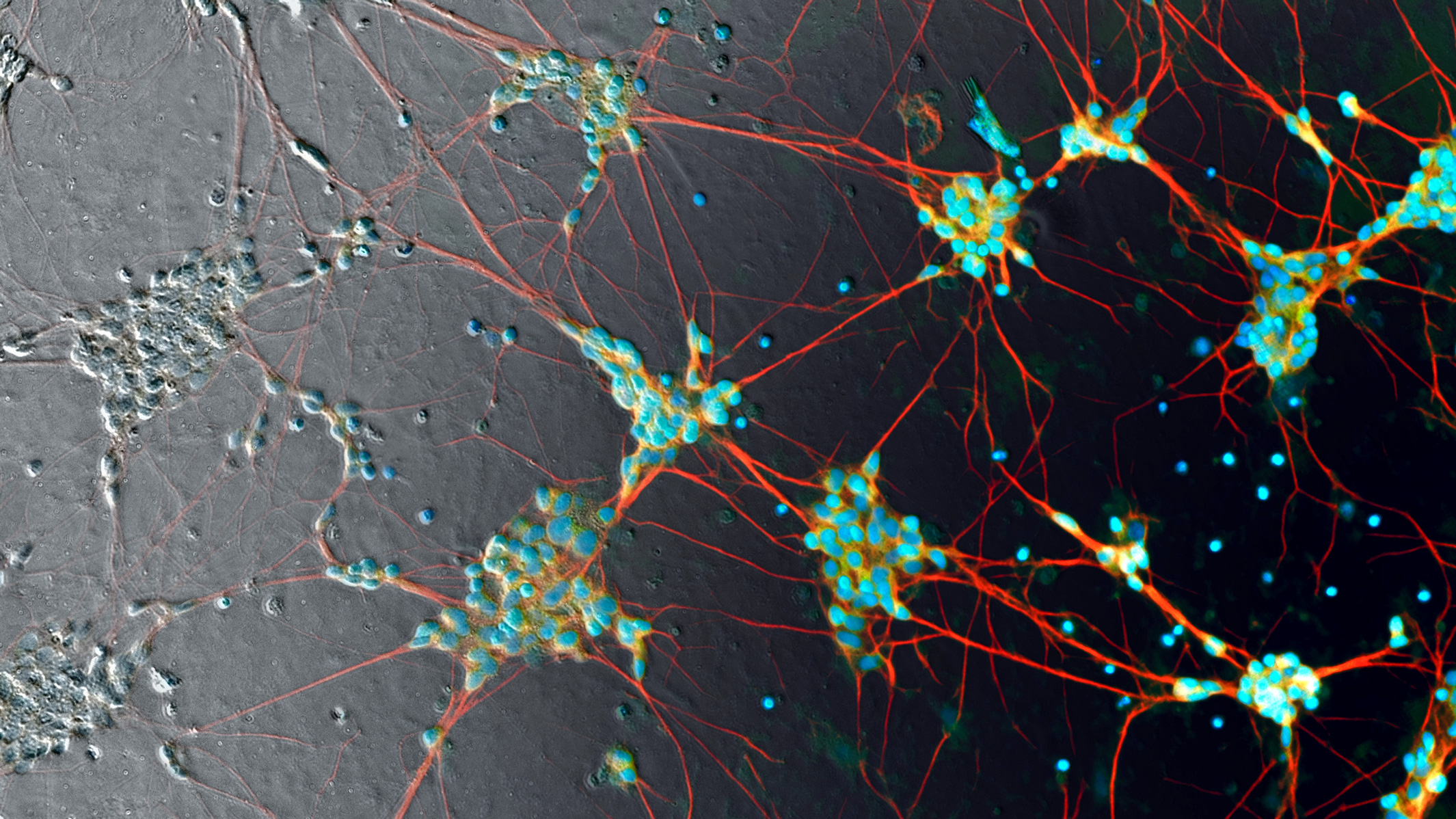
Human induced pluripotent stem cell neurons imaged in phase contrast (gray pixels, left) — currently processed manually with fluorescent labels (color pixels) to make them visible. That’s about to radically change. (credit: Google)
Researchers at Google, Harvard University, and Gladstone Institutes have developed and tested new deep-learning algorithms that can identify details in terabytes of bioimages, replacing slow, less-accurate manual labeling methods.
Deep learning is a type of machine learning that can analyze data, recognize patterns, and make predictions. A new deep-learning approach to biological images, which the researchers call “in silico labeling” (ISL), can automatically find and predict features in images of “unlabeled” cells (cells that have not been manually identified by using fluorescent chemicals).
The new deep-learning network can identify whether a cell is alive or dead, and get the answer right 98 percent of the time (humans can typically only identify a dead cell with 80 percent accuracy) — without requiring invasive fluorescent chemicals, which make it difficult to track tissues over time. The deep-learning network can also predict detailed features such as nuclei and cell type (such as neural or breast cancer tissue).
The deep-learning algorithms are expected to make it possible to handle the enormous 3–5 terabytes of data per day generated by Gladstone Institutes’ fully automated robotic microscope, which can track individual cells for up to several months.
The research was published in the April 12, 2018 issue of the journal Cell.
How to train a deep-learning neural network to predict the identity of cell features in microscope images

Using fluorescent labels with unlabeled images to train a deep neural network to bring out image detail. (Left) An unlabeled phase-contrast microscope transmitted-light image of rat cortex — the center image from the z-stack (vertical stack) of unlabeled images. (Right three images) Labeled images created with three different fluorescent labels, revealing invisible details of cell nuclei (blue), dendrites (green), and axons (red). The numbered outsets at the bottom show magnified views of marked subregions of images. (credit: Finkbeiner Lab)
To explore the new deep-learning approach, Steven Finkbeiner, MD, PhD, the director of the Center for Systems and Therapeutics at Gladstone Institutes in San Francisco, teamed up with computer scientists at Google.
“We trained the [deep learning] neural network by showing it two sets of matching images of the same cells: one unlabeled [such as the black and white "phase contrast"microscope image shown in the illustration] and one with fluorescent labels [such as the three colored images shown above],” explained Eric Christiansen, a software engineer at Google Accelerated Science and the study’s first author. “We repeated this process millions of times. Then, when we presented the network with an unlabeled image it had never seen, it could accurately predict where the fluorescent labels belong.” (Fluorescent labels are created by adding chemicals to tissue samples to help visualize details.)
The study used three cell types: human motor neurons derived from induced pluripotent stem cells, rat cortical cultures, and human breast cancer cells. For instance, the deep-learning neural network can identify a physical neuron within a mix of cells in a dish. It can go one step further and predict whether an extension of that neuron is an axon or dendrite (two different but similar-looking elements of the neural cell).
For this study, Google used TensorFlow, an open-source machine learning framework for deep learning originally developed by Google AI engineers. The code for this study, which is open-source on Github, is the result of a collaboration between Google Accelerated Science and two external labs: the Lee Rubin lab at Harvard and the Steven Finkbeiner lab at Gladstone.

Animation showing the same cells in transmitted light (black and white) and fluorescence (colored) imaging, along with predicted fluorescence labels from the in silico labeling model. Outset 2 shows the model predicts the correct labels despite the artifact in the transmitted-light input image. Outset 3 shows the model infers these processes are axons, possibly because of their distance from the nearest cells. Outset 4 shows the model sees the hard-to-see cell at the top, and correctly identifies the object at the left as DNA-free cell debris. (credit: Google)
Transforming biomedical research
“This is going to be transformative,” said Finkbeiner, who is also a professor of neurology and physiology at UC San Francisco. “Deep learning is going to fundamentally change the way we conduct biomedical science in the future, not only by accelerating discovery, but also by helping find treatments to address major unmet medical needs.”
In his laboratory, Finkbeiner is trying to find new ways to diagnose and treat neurodegenerative disorders, such as Alzheimer’s disease, Parkinson’s disease, and amyotrophic lateral sclerosis (ALS). “We still don’t understand the exact cause of the disease for 90 percent of these patients,” said Finkbeiner. “What’s more, we don’t even know if all patients have the same cause, or if we could classify the diseases into different types. Deep learning tools could help us find answers to these questions, which have huge implications on everything from how we study the disease to the way we conduct clinical trials.”
Without knowing the classifications of a disease, a drug could be tested on the wrong group of patients and seem ineffective, when it could actually work for different patients. With induced pluripotent stem cell technology, scientists could match patients’ own cells with their clinical information, and the deep network could find relationships between the two datasets to predict connections. This could help identify a subgroup of patients with similar cell features and match them to the appropriate therapy, Finkbeiner suggests.
The research was funded by Google, the National Institute of Neurological Disorders and Stroke of the National Institutes of Health, the Taube/Koret Center for Neurodegenerative Disease Research at Gladstone, the ALS Association’s Neuro Collaborative, and The Michael J. Fox Foundation for Parkinson’s Research.
Abstract of In Silico Labeling: Predicting Fluorescent Labels in Unlabeled Images
Microscopy is a central method in life sciences. Many popular methods, such as antibody labeling, are used to add physical fluorescent labels to specific cellular constituents. However, these approaches have significant drawbacks, including inconsistency; limitations in the number of simultaneous labels because of spectral overlap; and necessary pertur-bations of the experiment, such as fixing the cells, to generate the measurement. Here, we show that a computational machine-learning approach, which we call ‘‘in silico labeling’’ (ISL), reliably predicts some fluorescent labels from transmitted-light images of unlabeled fixed or live biological samples. ISL predicts a range of labels, such as those for nuclei, cell type (e.g., neural), and cell state (e.g., cell death). Because prediction happens in silico, the method is consistent, is not limited by spectral overlap, and does not disturb the experiment. ISL generates biological measurements that would otherwise be problematic or impossible to acquire.