Discovering new drugs and materials by ‘touching’ molecules in virtual reality
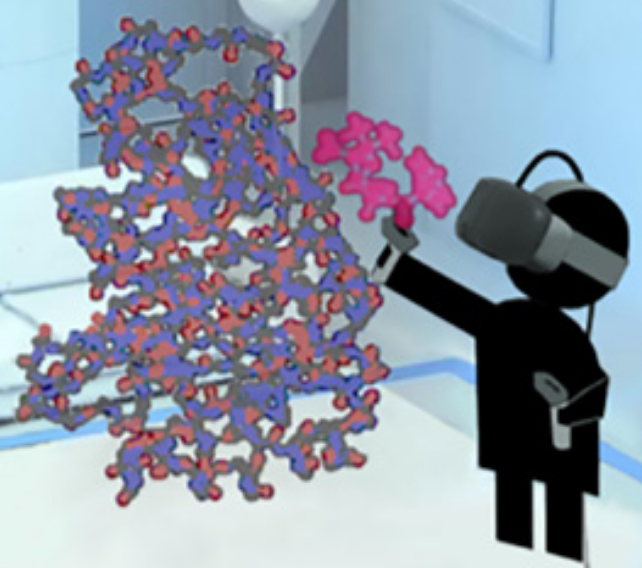
To figure out how to block a bacteria’s attempt to create multi-resistance to antibiotics, a researcher grabs a simulated ligand (binding molecule) — a type of penicillin called benzylpenicillin (red) — and interactively guides that molecule to dock within a larger enzyme molecule (blue-orange) called β-lactamase, which is produced by bacteria in an attempt to disable penicillin (making a patient resistant to a class of antibiotics called β-lactam). (credit: University of Bristol)
University of Bristol researchers, in collaboration with developers at Bristol based start-up Interactive Scientific, Oracle Corporation and a joint team of computer science and chemistry researchers, have designed and tested a new virtual reality (VR) cloud-based system intended to allow researchers to reach out and “touch” molecules as they move — folding them, knotting them, plucking them, and changing their shape to test how the molecules interact. The virtual reality cloud based system, called Nano Simbox, is the proprietary technology of Interactive Scientific, who collaborated with the University of Bristol to do the testing. Using an HTC Vive virtual-reality device, it could lead to creating new drugs and materials and improving the teaching of chemistry.
More broadly, the goal is to accelerate progress in nanoscale molecular engineering areas that include conformational mapping, drug development, synthetic biology, and catalyst design.
Real-time collaboration via the cloud

Two users passing a fullerene (C60) molecule back and forth in real time over a cloud-based network. The researchers are each wearing a VR head-mounted display (HMD) and holding two small wireless controllers that function as atomic “tweezers” to manipulate the real-time molecular dynamic of the C60 molecule. Each user’s position is determined using a real-time optical tracking system composed of synchronized infrared light sources, running locally on a GPU-accelerated computer. (credit: University of Bristol)
The multi-user system, developed by developed by a team led by University of Bristol chemists and computer scientists, uses an “interactive molecular dynamics virtual reality” (iMD VR) app that allows users to visualize and sample (with atomic-level precision) the structures and dynamics of complex molecular structures “on the fly” and to interact with other users in the same virtual environment.
Because each VR client has access to global position data of all other users, any user can see through his/her headset a co-located visual representation of all other users at the same time. So far, the system has uniquely allowed for simultaneously co-locating six users in the same room within the same simulation.
Testing on challenging molecular tasks
The team designed a series of molecular tasks for testing, using traditional mouse, keyboard, and touchscreens compared to virtual reality. The tasks included threading a small molecule through a nanotube, changing the screw-sense of a small organic helix, and tying a small string-like protein into a simple knot, and a variety of dynamic molecular problems, such as binding drugs to its target, protein folding, and chemical reactions. The researchers found that for complex 3D tasks, VR offers a significant advantage over current methods. For example, participants were ten times more likely to succeed in difficult tasks such as molecular knot tying.
Anyone can try out the tasks described in the open-access paper by downloading the software and launching their own cloud-hosted session.
David Glowacki | This video, made by University of Bristol PhD student Helen M. Deeks, shows the actions she took using a wireless set of “atomic tweezers” (using the HTC Vive) to interactively dock a single benzylpenicillin drug molecule into the active site of the β-lactamase enzyme.
David Glowacki | The video shows the cloud-mounted virtual reality framework, with several different views overlaid to give a sense of how the interaction works. The video outlines the four different parts of the user studies: (1) manipulation of buckminsterfullerene, enabling users to familarize themselves with the interactive controls; (2) threading a methane molecule through a nanotube; (3) changing the screw-sense of a helicene molecule; and (4) tying a trefoil knot in 17-Alanine.
Ref: Science Advances (open-access). Source: University of Bristol.
Research suggests that humans could one day regrow limbs
In the June 14, 2018, issue of the journal Cell, researchers at Stowers Institute for Medical Research published a landmark study whose findings have important implications for advancing the study of stem cell biology and regenerative medicine, according to the researchers.*
Over a century ago, scientists traced regenerative powers in a flatworm known as planaria to a special population of planaria adult stem cells called neoblasts (a type of adult pluripotent stem cell — meaning a cell that can transform into any type of cell). Scientists believe these neoblasts hold the secret to regeneration. But until recently, scientists lacked the tools necessary to identify exactly which of the individual types of neoblasts were actually capable of regeneration.
However, with a special technique that combined genomics, single-cell analysis, and imaging, the scientists were able to identify 12 different subgroups of neoblasts. The problem was to find the specific neoblasts that were pluripotent (able to create any kind of cell, instead of becoming specific cells, like muscle or skin). By further analyzing the 12 neoblast markers (genetic signatures), they narrowed it down one specific subgroup, called Nb2.

Planarian flatworm adult stem cells known as neoblasts can be clustered based on their gene expression profiles (left panel). A neoblast subpopulation termed Nb2 expresses the cell membrane protein TSPAN-1 (center panel, a representative Nb2 cell with TSPAN-1 protein shown in green and DNA in blue). Nb2 neoblasts were found to be able to repopulate stem cell-depleted animals (right panel, representative animals at different time points after Nb2 single-cell transplants). (credit: Stowers Institute for Medical Research)
To see if the Nb2 type of neoblast was truly capable of regeneration, they irradiated a group of planaria and then inserted the Nb2 into the planaria. They found the Nb2 subgroup was in fact able to repopulate the planaria.
“We have enriched for a pluripotent stem cell population, which opens the door to a number of experiments that were not possible before,” says senior author Alejandro Sánchez Alvarado, Ph.D. “The fact that the marker we discovered is expressed not only in planarians but also in humans suggests that there are some conserved mechanisms that we can exploit.
“I expect those first principles will be broadly applicable to any organism that ever relied on stem cells to become what they are today. And that essentially is everybody.”
Ref.: Cell. Source: Stowers Institute for Medical Research
* The work was funded by the Stowers Institute for Medical Research, the Howard Hughes Medical Institute, and the National Institute of General Medical Sciences of the National Institutes of Health.
Ultrasound-powered nanorobots clear bacteria and toxins from blood
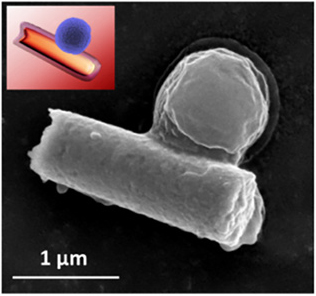
MRSA bacterium captured by a hybrid cell membrane-coated nanorobot (colored scanning electron microscope image and black and white image below) (credit: Esteban-Fernández de Ávila/Science Robotics)
Engineers at the University of California San Diego have developed tiny ultrasound-powered nanorobots that can swim through blood, removing harmful bacteria and the toxins they produce.
These proof-of-concept nanorobots could one day offer a safe and efficient way to detoxify and decontaminate biological threat agents — providing an fast alternative to the multiple, broad-spectrum antibiotics currently used to treat life-threatening pathogens like MRSA bacteria (an antibiotic-resistant staph strain). MRSA is considered a serious worldwide threat to public health.

The MRSA superbug (in yellow) is resistant to antibiotics and can lead to death (credit: National Institute of Allergy and Infectious Diseases)
Antimicrobial resistance (AMR) threatens the effective prevention and treatment of an ever-increasing range of infections caused by bacteria, parasites, viruses and fungi, according to the World Health Organization — an increasingly serious threat to global public health.
Trapping pathogens
The researchers coated gold nanowires with a hybrid of red blood cell membranes and platelets (tiny blood cells that help your body form clots to stop bleeding).*
- The platelets cloak the nanowires and attract bacterial pathogens, which become bound to the nanorobots.
- The red blood cells then absorb and neutralize the toxins produced by these bacteria.

Gold nanorobots coated in hybrid platelet/red blood cell membranes (colored scanning electron microscope image). (credit: Esteban-Fernández de Ávila/Science Robotics)
The interior gold nanowire body of the nanorobots responds to ultrasound, causing the nanorobots to swim around rapidly (no chemical fuel required) — mimicking the movement of natural motile cells (such as red blood cells). This mobility helps the nanorobots efficiently mix with their targets (bacteria and toxins) in blood and speed up detoxification.
The coating also protects the nanorobots from a process known as biofouling — when proteins collect onto the surface of foreign objects and prevent them from operating normally.
The nanorobots are just over one micrometer** (1,000 nanometers) long (for comparison, red blood cells have a diameter of 6 to 8 micrometers). The nanorobots can travel up to 35 micrometers per second in blood when powered by ultrasound.
In tests, the researchers used the nanorobots to treat blood samples contaminated with MRSA and their toxins. After five minutes, these blood samples had three times less bacteria and toxins than untreated samples.
Broad-spectrum detoxification
Future work includes tests in mice, making nanorobots out of biodegradable materials instead of gold, and tests of also using the nanorobots for drug delivery.
The ultimate research goal is not to use the nanorobots specifically for treating MRSA infections, but more generally for detoxifying biological fluids — “an important step toward the creation of a broad-spectrum detoxification robotic platform,” as the researchers note in a paper.
* The researchers created the nanorobots in three steps:
1. They created the hybrid coating by first separating entire membranes from platelets and red blood cells.
2. They applied ultrasound (high-frequency sound waves) to fuse the membranes together. (Since the membranes were taken from actual cells, they contain all their original-cell surface protein functions, including the ability of platelets to attract bacteria.)
3. They coated these hybrid membranes onto gold nanowires.
** A micrometer is one millionth of a meter, or one thousandth of a millimeter.
This work was supported by the Defense Threat Reduction Agency Joint Science and Technology Office for Chemical and Biological Defense.
Reference: Science Robotics. Source: UC San Diego.
Artificial sensory neurons may give future prosthetic devices and robots a subtle sense of touch

American and Korean researchers are creating an artificial nerve system for robots and humans. (credit: Kevin Craft)
Researchers at Stanford University and Seoul National University have developed an artificial sensory nerve system that’s a step toward artificial skin for prosthetic limbs, restoring sensation to amputees, and giving robots human-like reflexes.*
Their rudimentary artificial nerve circuit integrates three previously developed components: a touch-pressure sensor, a flexible electronic neuron, and an artificial synaptic transistor modeled on human synapses.
Here’s how the artificial nerve circuit works:

(Biological model) Pressures applied to afferent (sensory) mechanoreceptors (pressure sensors, in this case) in the finger change the receptor potential (voltage) of each mechanoreceptor. The receptor potential changes combine and initiate action potentials in the nerve fiber, connected to a heminode in the chest. The nerve fiber forms synapses with interneurons in the spinal cord. Action potentials from multiple nerve fibers combine through the synapses and contribute to information processing (via postsynaptic potentials). (credit: (Yeongin Kim (Stanford University), Alex Chortos(Stanford University), Wentao Xu (Seoul National University), Zhenan Bao (Stanford University), Tae-Woo Lee (Seoul National University))
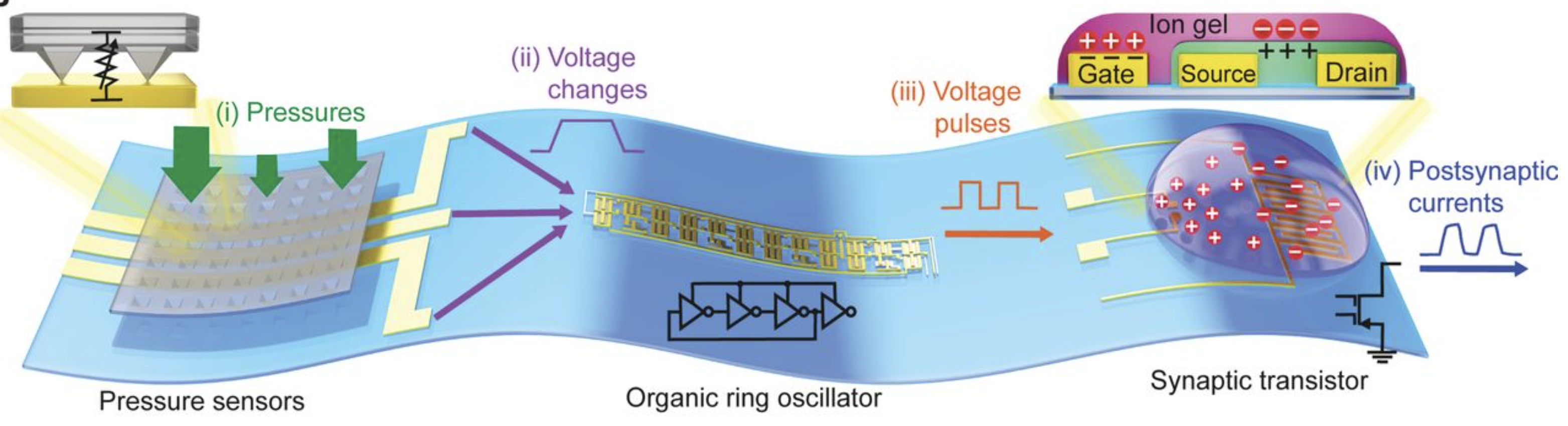
(Artificial model) Illustration of a corresponding artificial afferent nerve system made of pressure sensors, an organic ring oscillator (simulates a neuron), and a transistor that simulates a synapse. (Only one ring oscillator connected to a synaptic transistor is shown here for simplicity.) Colors of parts match corresponding colors in the biological version. (credit: Yeongin Kim (Stanford University), Alex Chortos (Stanford University), Wentao Xu (Seoul National University), Zhenan Bao (Stanford University), Tae-Woo Lee (Seoul National University))

(Photo) Artificial sensor, artificial neuron, and artificial synapse. (credit: Yeongin Kim (Stanford University), Alex Chortos(Stanford University), Wentao Xu (Seoul National University), Zhenan Bao (Stanford University), Tae-Woo Lee (Seoul National University))
Experiments with the artificial nerve circuit
In a demonstration experiment, the researchers used the artificial nerve circuit to activate the twitch reflex in the knee of a cockroach.

A cockroach insect (A) with an attached artificial mechanosensory nerve was used in this experiment. The artificial afferent nerve (B) was connected to the biological motor (movement) nerves of a detached insect leg (B, lower right) to demonstrate a hybrid reflex arc (such as a knee reflex). Applied pressure caused a reflex movement of the leg. A force gauge (C) was used to measure the force of the reflex movements of the disabled insect leg. (credit: Yeongin Kim (Stanford University), Alex Chortos(Stanford University), Wentao Xu (Seoul National University), Zhenan Bao (Stanford University), Tae-Woo Lee (Seoul National University))
The researchers did another experiment that showed how the artificial nerve system could be used to identify letters in the Braille alphabet.
Improving robot and human sensory abilites
The researchers “used a knee reflex as an example of how more-advanced artificial nerve circuits might one day be part of an artificial skin that would give prosthetic devices or robots both senses and reflexes,” noted Chiara Bartolozzi, Ph.D., of Istituto Italiano Di Tecnologia, writing in a Science commentary on the research.
Tactile information from artificial tactile systems “can improve the interaction of a robot with objects,” says Bartolozzi, who is involved in research with the iCub robot.
“In this scenario, objects can be better recognized because touch complements the information gathered from vision about the shape of occluded or badly illuminated regions of the object, such as its texture or hardness. Tactile information also allows objects to be better manipulated — for example, by exploiting contact and slip detection to maintain a stable but gentle grasp of fragile or soft objects (see the photo). …
“Information about shape, softness, slip, and contact forces also greatly improves the usability of upper-limb prosthetics in fine manipulation. … The advantage of the technology devised by Kim et al. is the possibility of covering at a reasonable cost larger surfaces, such as fingers, palms, and the rest of the prosthetic device.
“Safety is enhanced when sensing contacts inform the wearer that the limb is encountering obstacles. The acceptability of the artificial hand by the wearer is also improved because the limb is perceived as part of the body, rather than as an external device. Lower-limb prostheses can take advantage of the same technology, which can also provide feedback about the distribution of the forces at the foot while walking.”
Next research steps
The researchers plan next to create artificial skin coverings for prosthetic devices, which will require new devices to detect heat and other sensations, the ability to embed them into flexible circuits, and then a way to interface all of this to the brain. They also hope to create low-power, artificial sensor nets to cover robots. The idea is to make them more agile by providing some of the same feedback that humans derive from their skin.
“We take skin for granted but it’s a complex sensing, signaling and decision-making system,” said Zhenan Bao, Ph.D., a Stanford professor of chemical engineering and one of the senior authors. “This artificial sensory nerve system is a step toward making skin-like sensory neural networks for all sorts of applications.”
This milestone is part of Bao’s quest to mimic how skin can stretch, repair itself, and, most remarkably, act like a smart sensory network that knows not only how to transmit pleasant sensations to the brain, but also when to order the muscles to react reflexively to make prompt decisions.
The synaptic transistor is the brainchild of Tae-Woo Lee of Seoul National University, who spent his sabbatical year in Bao’s Stanford lab to initiate the collaborative work.
Reference: Science May 31. Source: Stanford University and Seoul National University.
* This work was funded by the Ministry of Science and ICT, Korea; by Seoul National University (SNU); by Samsung Electronics; by the National Nanotechnology Coordinated Infrastructure; and by the Stanford Nano Shared Facilities (SNSF). Patents related to this work are planned.
Ingestible capsule uses light-emitting bacteria to monitor gastrointestinal health

MIT-designed biosensor capsule uses genetically engineered light-emitting bacteria (right) to detect molecules that identify bleeding or other gastrointestinal problems. Ultra-low-power electronics (left) sense the light and send diagnostic information wirelessly to a cellphone. (credit: Lillie Paquette/MIT)
MIT engineers have designed and built a tiny ingestible biosensor* capsule that can diagnose gastrointestinal problems, and the engineers demonstrated its ability to detect bleeding in pigs.
Currently, if patients are suspected to be bleeding from a gastric ulcer, for example, they have to undergo an endoscopy to diagnose the problem, which often requires the patient to be sedated.
If the engineers can shrink the sensor capsule and detect a variety of other conditions, the research could potentially transform the diagnosis of gastrointestinal diseases and conditions, according to the researchers.
Diagnosing gastrointestinal diseases in real time

To detect diseases or conditions, the genetically engineered bacteria (green) are placed into multiple wells (blue), covered by a semipermeable membrane (white) that allows small molecules (red) from the surrounding environment to diffuse through. The bacteria luminesce (glow) when they sense the specific type of molecule they are designed for. (In the experiment with pigs, heme — part of the red hemoglobin blood pigment — indicated bleeding.) A phototransistor (brown) measures the amount of light produced by the bacterial cells and relays that information to a microprocessor in the capsule, which then sends a wireless signal to a nearby computer or smartphone. (credit: Mark Mimee et al./Science)
The researchers showed that the ingestible biosensor could correctly determine whether any blood was present in the pig’s stomach. They anticipate that this type of sensor could be deployed for either one-time use or to remain in the digestive tract for several days or weeks, sending continuous signals. The sensors could also be designed to carry multiple strains of bacteria, allowing for diagnosing multiple diseases and conditions.
The researchers plan to reduce the size of the sensor capsule (currently 10 millimeters wide by 30 millimeters long) and to study how long the bacteria cells can survive in the digestive tract. They also hope to develop sensors for gastrointestinal conditions other than bleeding.**
Reference: Science. Source: MIT.
* The sensor requires only 13 microwatts of power. The researchers equipped the sensor with a 2.7-volt battery, which they estimate could power the device for about 1.5 months of continuous use. They say it could also be powered by a voltaic cell sustained by acidic fluids in the stomach, using previously developed MIT technology.
** For example, one of the sensors they designed detects a sulfur-containing ion called thiosulfate, which is linked to inflammation and could be used to monitor patients with Crohn’s disease or other inflammatory conditions. Another one detects a bacterial signaling molecule called AHL, which can serve as a marker for gastrointestinal infections because different types of bacteria produce slightly different versions of the molecule.
MIT’s modular plug-and-play blocks allow for building medical diagnostic devices

Tiny 1/2-inch, low-cost “Ampli blocks” can be assembled to create diagnostic devices. The blocks, which simply consist of a tiny sheet of paper or glass fiber sandwiched between a plastic or metal block and a glass cover, snap together to form a complete diagnostic procedure. Some of the blocks contain channels for samples to flow straight through, some have turns, and some can receive a sample from a pipette, or mix multiple reagents (chemicals) together. The blocks are color-coded by function, making it easy to assemble pre-designed devices (the researchers plan to put instructions online). (credit: MIT Little Devices Lab)
Researchers at MIT’s Little Devices Lab have developed a set of modular “plug-and-play” blocks that can be put together in different ways to produce medical diagnostic devices for detecting cancer and infectious diseases such as Zika virus.
The “Ampli blocks” require little expertise to assemble, and can test blood glucose levels in diabetic patients or detect viral infection, for example. They are inexpensive (about 6 U.S. cents for four blocks) and no refrigeration is required, making them particularly important for small, low-resources laboratories in the developing world. Small labs can now create their own libraries of plug-and-play diagnostics to independently treat their own local patient populations.
Customized diagnostics on a modular “biochemical breadboard”
KurzweilAI has previously reported on small, portable diagnostic devices based on chemical reactions that occur on paper strips. But these devices haven’t been widely deployed. That’s mainly because they’re not designed for mass-producing a diagnostic test — especially when a disease doesn’t affect a large number of people, the researchers say.
The Little Devices Lab has created a kit of DIY modular components that can be easily put together to generate exactly what a lab needs. So far, the MIT lab has created about 40 different building blocks that lab workers around the world could easily assemble on their own. This approach is similar to assembling radios and other electronic devices from commercially available electronic “breadboards” in the 1970s, or more recently, creating your own homemade computer gadgets with an Arduino breadboard or a Raspberry Pi computer.

Jose Gomez-Marquez, co-director of MIT’s Little Devices Lab, holds a sheet of tiny diagnostic papers, which can be easily printed and attached to blocks to form diagnostic devices. (credit:Melanie Gonick/MIT)
The reusable blocks can also perform different biochemical functions. Many contain antibodies that can detect a specific molecule in a blood or urine sample. The antibodies are attached to nanoparticles that change color when the target molecule is present, indicating a positive result.
These blocks can be aligned in different ways, allowing the user to create diagnostics based on one reaction or a series of reactions. In one example, the researchers combined blocks that detect three different molecules to create a test for isonicotinic acid, which can reveal whether tuberculosis patients are taking their medication.
The researchers also showed that these blocks can outperform previous versions of paper diagnostic devices in some cases. For example, they found that they could run a sample back and forth over a test strip multiple times, enhancing the signal. This could make it easier to get reliable results from urine and saliva samples, which are usually more dilute than blood samples, but are easier to obtain from patients.
Getting the technology into the hands of small labs and non-experts
The MIT team is now working on tests for human papilloma virus, malaria, and Lyme disease, among others. They are also working on blocks that can synthesize useful compounds, including drugs, and even on blocks that incorporate electrical components such as LEDs.
The ultimate goal is to get the technology into the hands of small labs (and non-experts) in both industrialized and developing countries, so they can create their own diagnostics. The MIT team has already sent Ampli-block kits to labs in Chile and Nicaragua, where they have been used to develop devices to monitor patient adherence to TB treatment and to test for a genetic variant that makes malaria more difficult to treat.
The MIT researchers are now investigating large-scale manufacturing techniques. They hope to launch a company to manufacture and distribute the kits around the world. They’re also opening up the platform to other researchers.
Reference: Advanced Healthcare Materials May 16, 2018. Source: MIT May 16, 2018.
Three dramatic new ways to visualize brain tissue and neuron circuits

Visualizing the brain: Here, tissue from a human dentate gyrus (a part of the brain’s hippocampus that is involved in the formation of new memories) was imaged transparently in 3D and colored-coded to reveal the distribution and types of nerve cells. (credit: The University of Hong Kong)
Visualizing human brain tissue in vibrant transparent colors
Neuroscientists from The University of Hong Kong (HKU) and Imperial College London have developed a new method called “OPTIClear” for 3D transparent color visualization (at the microscopic level) of complex human brain circuits.
To understand how the brain works, neuroscientists map how neurons (nerve cells) are wired to form circuits in both healthy and disease states. To do that, the scientists typically cut brain tissues into thin slices. Then they trace the entangled fibers across those slices — a complex, laborious process.
Making human tissues transparent. OPTIClear replaces that process by “clearing” (making tissues transparent) and using fluorescent staining to identify different types of neurons. In one study of more than 3,000 large neurons in the human basal forebrain, the researchers were able to reduce the time from about three weeks to five days to visualize neurons, glial cells, and blood vessels in exquisite 3D detail. Previous clearing methods (such as CLARITY) have been limited to rodent tissue.
Reference (open access): Nature Communications March 14, 2018. Source: HKU and Imperial College London, May 7, 2018
Watching millions of brain cells in a moving animal for the first time

Neurons in the hippocampus flash on and off as a mouse walks around with tiny camera lenses on its head. (credit: The Rockefeller University)
It’s a neuroscientist’s dream: being able to track the millions of interactions among brain cells in animals that move about freely — allowing for studying brain disorders. Now a new invention, developed at The Rockefeller University and reported today, is expected to give researchers a dynamic tool to do that just that, eventually in humans.
The new tool can track neurons located at different depths within a volume of brain tissue in a freely moving rodent, or record the interplay among neurons when two animals meet and interact socially.
Microlens array for 3D recording. The technology consists of a tiny microscope attached to a mouse’s head, with a group of lenses called a “microlens array.” These lenses enable the microscope to capture images from multiple angles and depths on a sensor chip, producing a three-dimensional record of neurons blinking on and off as they communicate with each other through electrochemical impulses. (The mouse neurons are genetically modified to light up when they become activated.) A cable attached to the top of the microscope transmits the data for recording.
One challenge: Brain tissue is opaque, making light scatter, which makes it difficult to pinpoint the source of each neuronal light flash. The researchers’ solution: a new computer algorithm (program), known as SID, that extracts additional information from the scattered emission light.
Reference: Nature Methods. Source: The Rockefeller University May 7, 2018
Brain cells interacting in real time
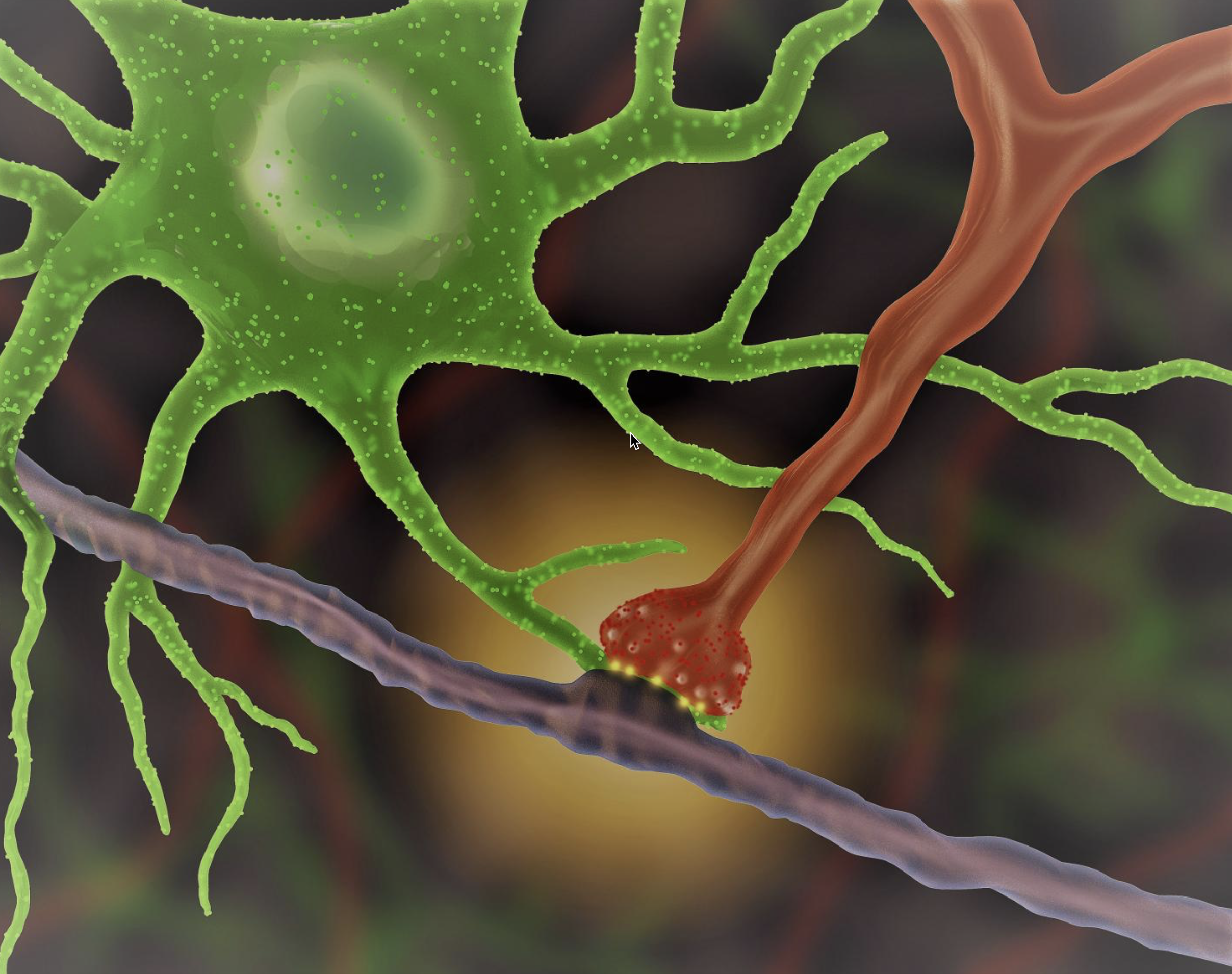
Illustration: An astrocyte (green) interacts with a synapse (red), producing an optical signal (yellow). (credit: UCLA/Khakh lab)
Researchers at the David Geffen School of Medicine at UCLA can now peer deep inside a mouse’s brain to watch how star-shaped astrocytes (support glial cells in the brain) interact with synapses (the junctions between neurons) to signal each other and convey messages.
The method uses different colors of light that pass through a lens to magnify objects that are invisible to the naked eye. The viewable objects are now far smaller than those viewable by earlier techniques. That enables researchers to observe how brain damage alters the way astrocytes interact with neurons, and develop strategies to address these changes, for example.
Astrocytes are believed to play a key role in neurological disorders like Lou Gehrig’s, Alzheimer’s, and Huntington’s disease.
Reference: Neuron. Source: UCLA Khakh lab April 4, 2018.
round-up | Hawking’s radical instant-universe-as-hologram theory and the scary future of information warfare
Stephen Hawking’s final cosmology theory says the universe was created instantly (no inflation, no singularity) and it’s a hologram
There was no singularity just after the big bang (and thus, no eternal inflation) — the universe was created instantly. And there were only three dimensions. So there’s only one finite universe, not a fractal or a multiverse — and we’re living in a projected hologram. That’s what Hawking and co-author Thomas Hertog (a theoretical physicist at the Catholic University of Leuven) have concluded — contradicting Hawking’s former big-bang singularity theory (with time as a dimension).
Problem: So how does time finally emerge? “There’s a lot of work to be done,” admits Hertog. Citation (open access): Journal of High Energy Physics, May 2, 2018. Source (open access): Science, May 2, 2018
Molecular movies of RNA guide drug discovery — a new paradigm for drug discovery
Duke University scientists have invented a technique that combines nuclear magnetic resonance imaging and computationally generated movies to capture the rapidly changing states of an RNA molecule.
It could lead to new drug targets and allow for screening millions of potential drug candidates. So far, the technique has predicted 78 compounds (and their preferred molecular shapes) with anti-HIV activity, out of 100,000 candidate compounds. Citation: Nature Structural and Molecular Biology, May 4, 2018. Source: Duke University, May 4, 2018.

Chromium tri-iodide magnetic layers between graphene conductors. By using four layers, the storage density could be multiplied. (credit: Tiancheng Song)
Atomically thin magnetic memory
University of Washington scientists have developed the first 2D (in a flat plane) atomically thin magnetic memory — encoding information using magnets that are just a few layers of atoms in thickness — a miniaturized, high-efficiency alternative to current disk-drive materials.
In an experiment, the researchers sandwiched two atomic layers of chromium tri-iodide (CrI3) — acting as memory bits — between graphene contacts and measured the on/off electron flow through the atomic layers.
The U.S. Dept. of Energy-funded research could dramatically increase future data-storage density while reducing energy consumption by orders of magnitude. Citation: Science, May 3, 2018. Source: University of Washington, May 3, 2018.

Definitions of artificial intelligence (credit: House of Lords Select Committee on Artificial Intelligence)
A Magna Carta for the AI age
A report by the House of Lords Select Committee on Artificial Intelligence in the U.K. lays out “an overall charter for AI that can frame practical interventions by governments and other public agencies.”
The key elements: Be developed for the common good. Operate on principles of intelligibility and fairness: users must be able to easily understand the terms under which their personal data will be used. Respect rights to privacy. Be grounded in far-reaching changes to education. Teaching needs reform to utilize digital resources, and students must learn not only digital skills but also how to develop a critical perspective online. Never be given the autonomous power to hurt, destroy or deceive human beings.
Source: The Washington Post, May 2, 2018.
The future of information warfare
Memes and social networks have become weaponized, but many governments seem ill-equipped to understand the new reality of information warfare.
The weapons include: Computational propaganda: digitizing the manipulation of public opinion; advanced digital deception technologies; malicious AI impersonating and manipulating people; and AI-generated fake video and audio. Counter-weapons include: Spotting AI-generated people; uncovering hidden metadata to authenticate images and videos; blockchain for tracing digital content back to the source; and detecting image and video manipulation at scale.
Source (open-access): CB Insights Research Brief, May 3, 2018.
New immunotherapy treatment for lung cancer dramatically improves survival, researchers report
An immunotherapy treatment — one that boosts the immune system — has improved survival in people newly diagnosed with the most common form of lung cancer (advanced non–small-cell lung cancer), according to an open-access study published in the New England Journal of Medicine.
The study results were presented last Monday, April 16, at the annual American Association for Cancer Research conference in Chicago.
Cutting the risk of dying in half. The new study, led by thoracic medical oncologist Leena Gandhi, MD, PhD, associate professor of medicine and director of the thoracic medical oncology program at NYU’s Perlmutter Cancer Center, shows that treating lung cancer by a combination of immunotherapy with Merck’s Keytruda (aka pembrolizumab) and chemotherapy is more effective than chemotherapy alone, according to a statement by NYU Langone Health.
The combination cut in half the risk of dying or having the cancer worsen, compared to chemo alone, after nearly one year, the Associated Press reported in The New York Times. “The results are expected to quickly set a new standard of care for about 70,000 patients each year in the United States whose lung cancer has already spread by the time it’s found,” the AP stated.
“Another study found that an immunotherapy combo — the Bristol-Myers Squibb drugs Opdivo and Yervoy — worked better than chemo for delaying the time until cancer worsened in advanced lung cancer patients whose tumors have many gene flaws, as nearly half do. But the benefit lasted less than two months on average and it’s too soon to know if the combo improves overall survival, as Keytruda did.”
Removing a cloak. All three of these “checkpoint inhibitor” treatments remove a “cloak” that some cancer cells have that hides the cancer cells from the immune system.
These immune-therapy treatments — which are administered through IVs and cost about $12,500 a month — worked for only about half of patients. But that’s far better than chemo alone has done in the past, notes the AP.
The American Cancer Society estimates that in 2018, there will be about 234,030 new cases of lung cancer in the U.S and about 154,050 deaths from lung cancer.