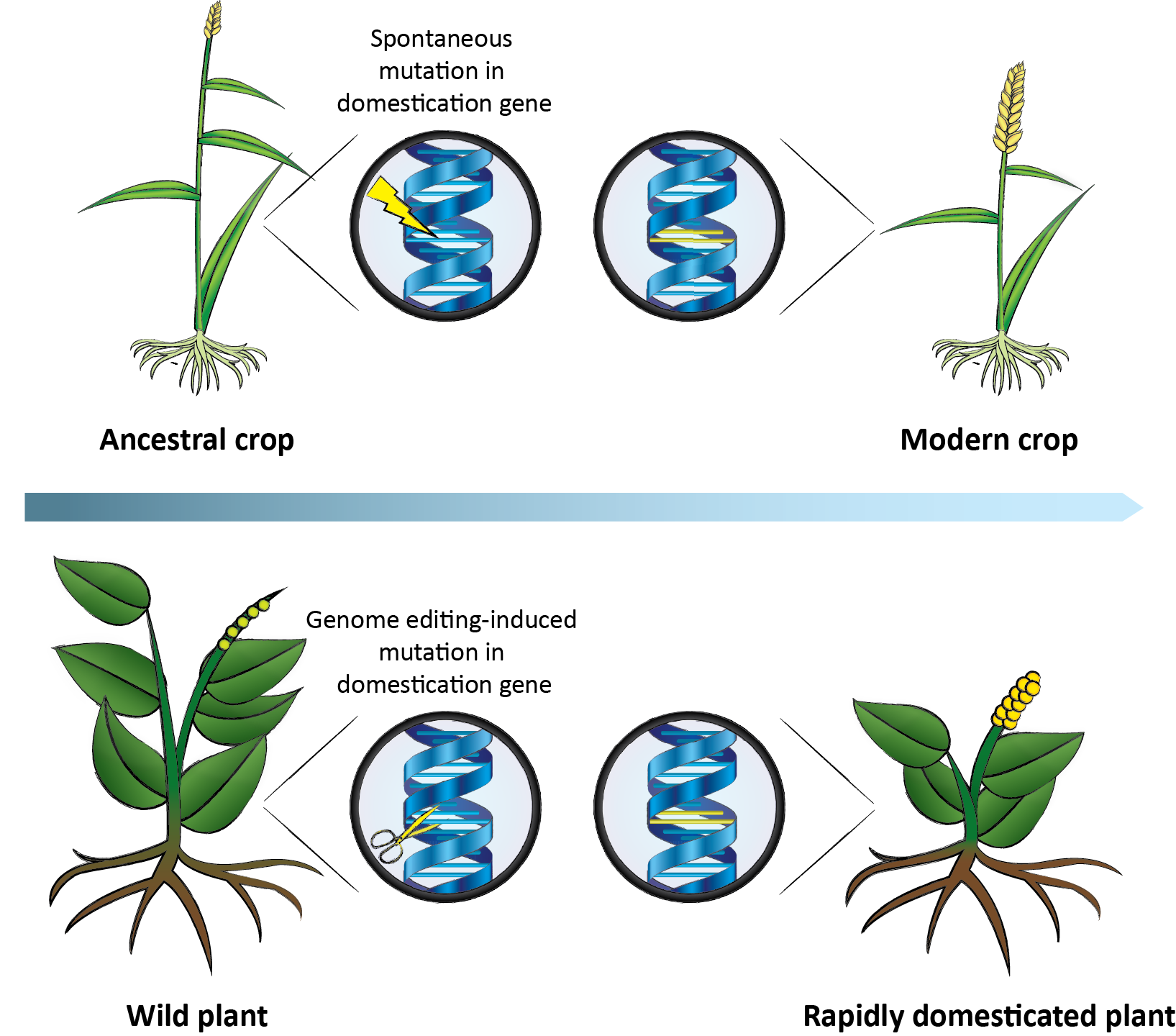
Accelerating the domestication of wild plants. During the domestication of ancestral crops, plants carrying spontaneous mutations in domestication genes were selected for. The same genes can be targeted in wild plants by genome editing, resulting in a rapidly domesticated plant. (credit: Cell)
Here’s a radical new idea for creating new GMO (genetically modified organism) plants that may appeal to staunch organic-food consumers/farmers and even #NonGMOProjectVerified advocates: don’t insert a foreign gene in today’s domestic plants — delete already existing genes in semi-domesticated or even wild plants to make those plants more domestic, and reducing pesticide use in the process.
“All of the plants we eat today are mutants, but the crops we have now were selected for over thousands of years, and their mutations … such as reduced bitterness and those that facilitate easy harvest … arose by chance,” says Michael Palmgren, a botanist who heads an interdisciplinary think tank* called “Plants for a Changing World” at the University of Copenhagen. “With gene editing, we can create ‘biologically inspired organisms’ in that we don’t want to improve nature, we want to benefit from what nature has already created.”
Palmgen is senior author of an open-access review published March 2 in the journal Trends in Plant Science.
How to turn nitrogen in the atmosphere into fertilizer, reducing environmental damage
This strategy could also address problems from pesticide use and the damaging impact of large-scale agriculture on the environment. For example, runoff from excess nitrogen in fertilizers is a common pollutant; however, wild legumes, through symbiosis with bacteria, can turn nitrogen available in the atmosphere into their own fertilizer, he suggests.

Future logo? (credit: KurzweilAI)
Out of the more than 300,000 plant species in existence, fewer than 200 are commercially important, and only three species — rice, wheat, and maize — account for most of the plant matter that humans consume, partly because in the history of agriculture, mutations arose that made these crops the easiest to harvest, the reseachers note.
But with CRISPR technology, we don’t have to wait for nature to help us domesticate plants, argue the researchers. Instead, gene editing could make, for example, wild legumes, quinoa, or amaranth, which are already sustainable and nutritious, more farmable.
The approach has already been successful in accelerating domestication of undervalued crops using less precise gene-editing methods. For example, researchers used chemical mutagenesis to induce random mutations in weeping rice grass, an Australian wild relative of domestic rice, to make it more likely to hold onto its seeds after ripening. And in wild field cress, a type of weedy grass, scientists silenced genes with RNA interference involved with fatty acid synthesis, resulting in improved seed oil quality.
Palmgren’s group published a related open-access paper two years ago on using gene editing to make domesticated plants more “wild” and thus hardier for organic farmers.
While we’re at it, what about pharming (creating pharmaceuticals from plants) — using genetically modified wild plants?
* Supported by the University of Copenhagen Excellence Programme for Interdisciplinary Research.
Abstract of Accelerating the Domestication of New Crops: Feasibility and Approaches
The domestication of new crops would promote agricultural diversity and could provide a solution to many of the problems associated with intensive agriculture. We suggest here that genome editing can be used as a new tool by breeders to accelerate the domestication of semi-domesticated or even wild plants, building a more varied foundation for the sustainable provision of food and fodder in the future. We examine the feasibility of such plants from biological, social, ethical, economic, and legal perspectives.