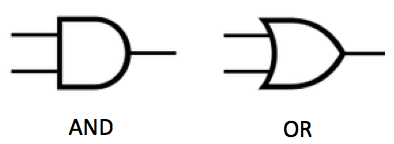
MIT researchers have developed synthetic biological circuits (from bacteria, for example, as shown here) that combine analog and digital computation as “living therapeutics” to treat major diseases and rare genetic disorders (credit: Synlogic)
MIT researchers have developed synthetic biological circuits that combine both analog (continuous) and digital (discrete) computation — allowing living cells to carry out complex processing operations, such as releasing a drug in response to low glucose levels.
The research is presented in an open-access paper published in the journal Nature Communications.
Background: analog vs. digital biological circuits
Like electronic circuits, living cells are capable of performing computations that are either continuous (analog) — like the way eyes adjust to gradual changes in the light levels — or digital, involving simple discrete on or off processes, such as a cell’s self-programmed death (apoptosis). Current synthetic biological systems, in contrast, have tended to focus on either analog or digital processing, limiting the range of uses.
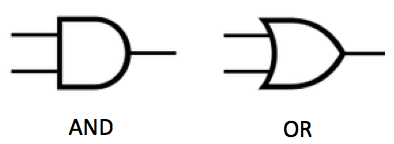
Two basic logic circuits. An AND gate fires only if both inputs are “true” (for example, both inputs have a 1 volt signal, not zero). An OR gate fires if either (or both) of the inputs is true (for example, the top input has a 1 volt signal and the bottom input has zero. (credit: KurzweilAI)
Digital systems are based on a simple binary output, such as 0 or 1, so performing complex computational operations requires the use of a large number of parts (such as AND and OR logic gates) to make the decision, which is difficult to achieve in synthetic biological systems. (There are seven basic logic gates: AND, OR, XOR, NOT, NAND, NOR, and XNOR, as explained here.)
Using genes (instead of voltages), synthetic biologists design genetic circuits (arrangements of DNA components) that can perform new functions. For example, here’s a gene circuit that was constructed using Escherichia coli bacteria (source: Imperial College London/Nature Communications study):
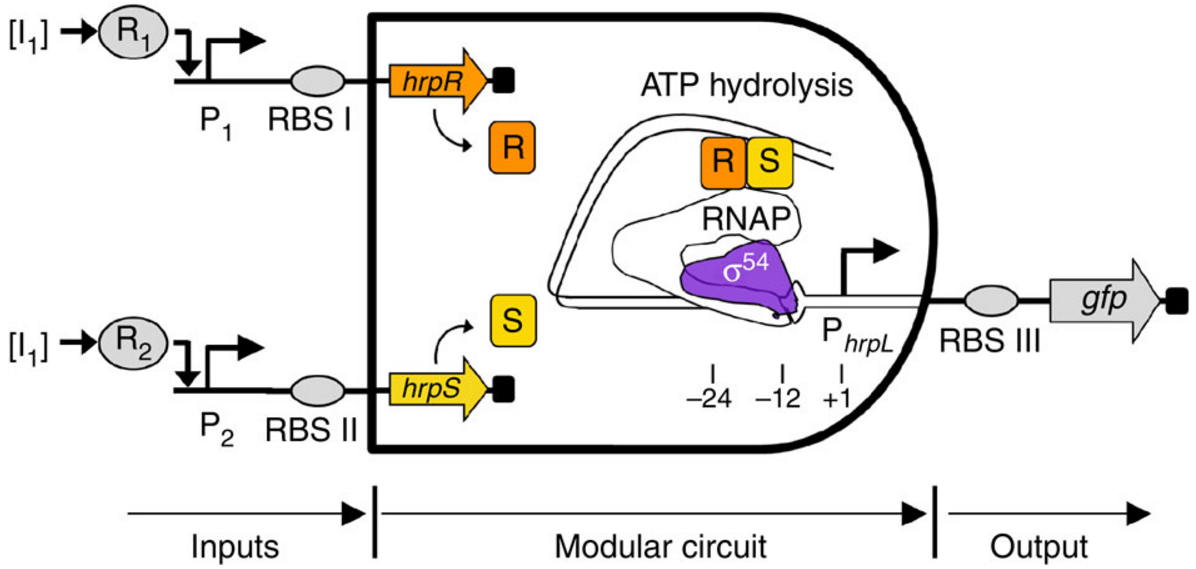
Example of a biological AND gate. Two environment-responsive gene promoters (a region of DNA that initiates transcription — that is, copying a particular segment of DNA into RNA), P1 and P2, act as the inputs to drive the transcriptions of hrpR and hrpS genes, and respond to small molecules. Transcription of the output promoter gfp is turned on only when both proteins HrpR and HrpS are present. (credit: Baojun Wang et al./Nature Communications)
“Most of the work in synthetic biology has focused on the digital approach, because [digital systems] are much easier to program,” says Timothy Lu, an associate professor of electrical engineering and computer science and of biological engineering, and head of the Synthetic Biology Group at MIT’s Research Laboratory of Electronics.
The new synthetic circuits can measure the level of an analog input, such as a particular chemical relevant to a disease, and then make a binary decision — for example, turning on an output, such as a drug that treats the disease if the level is in the right range.
The new circuits are based on multiple elements. For example, a threshold module consists of a sensor that detects analog levels of a particular chemical, which controls the expression of the second digital component, a recombinase gene, which can then switch on or off a segment of DNA by converting it into a digital (on or off) output. (This conversion process is similar to electronic devices known as comparators, which take analog input signals and convert them into a digital output.)
If the concentration of the chemical reaches a certain level, the threshold module expresses the recombinase gene, causing it to flip the DNA segment (which contains a gene or gene-regulatory element, which then alters the expression of a desired output).
“So this is how we take an analog input, such as a concentration of a chemical, and convert it into a 0 or 1 signal,” Lu says. “And once that is done, and you have a piece of DNA that can be flipped upside down, then you can put together any of those pieces of DNA to perform digital computing,” he says.
Ternary logic for three-way glucose decisions
The team has also built an analog-to-digital converter circuit that implements ternary (three-valued) logic. The circuit, which is capable of producing two different outputs, will only switch on in response to either a high or low concentration range of an input.
In the future, the circuit could be used to detect glucose levels in the blood and respond in one of three ways depending on the concentration, he says. “If the glucose level was too high, you might want your cells to produce insulin, if the glucose was too low you might want them to make glucagon, and if it was in the middle you wouldn’t want them to do anything,” he says.
Similar analog-to-digital converter circuits could also be used to detect a variety of chemicals, simply by changing the sensor, Lu says.
Detecting inflammation and environmental conditions
The researchers are investigating the idea of using analog-to-digital converters to detect levels of inflammation in the gut caused by inflammatory bowel disease, for example, and releasing different amounts of a drug in response.
Immune cells used in cancer treatment could also be engineered to detect different environmental inputs, such as oxygen or tumor lysis (cell breakdown) levels, and vary the immune-call therapeutic activity in response.
Other research groups are also interested in using the devices for environmental applications, such as engineering cells that detect concentrations of water pollutants, Lu says.
The research team recently created a spinout company, called Synlogic, which is now attempting to use simple versions of the circuits to engineer probiotic bacteria that can treat diseases in the gut. The company hopes to begin clinical trials of these bacteria-based treatments within the next 12 months.
Abstract of Synthetic mixed-signal computation in living cells
Living cells implement complex computations on the continuous environmental signals that they encounter. These computations involve both analogue- and digital-like processing of signals to give rise to complex developmental programs, context-dependent behaviours and homeostatic activities. In contrast to natural biological systems, synthetic biological systems have largely focused on either digital or analogue computation separately. Here we integrate analogue and digital computation to implement complex hybrid synthetic genetic programs in living cells. We present a framework for building comparator gene circuits to digitize analogue inputs based on different thresholds. We then demonstrate that comparators can be predictably composed together to build band-pass filters, ternary logic systems and multi-level analogue-to-digital converters. In addition, we interface these analogue-to-digital circuits with other digital gene circuits to enable concentration-dependent logic. We expect that this hybrid computational paradigm will enable new industrial, diagnostic and therapeutic applications with engineered cells.