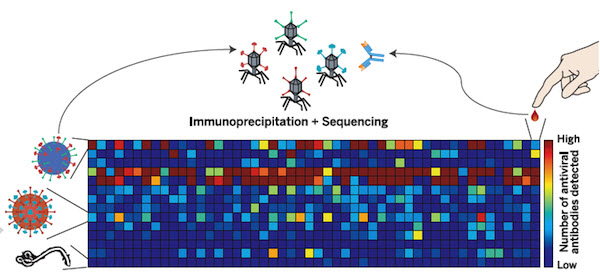
Systematic viral epitope scanning (VirScan). This method allows comprehensive analysis of antiviral antibodies in human sera. VirScan combines DNA microarray synthesis and bacteriophage display to create a uniform, synthetic representation of peptide epitopes comprising the human virome. Immunoprecipitation and high-throughput DNA sequencing reveal the peptides recognized by antibodies in the sample. The color of each cell in the heatmap depicts the relative number of antigenic epitopes detected for a virus (rows) in each sample (columns). (credit: George J. Xu et al./Science).
New technology called called VirScan developed by Howard Hughes Medical Institute (HHMI) researchers makes it possible to test for current and past infections with any known human virus by analyzing a single drop of a person’s blood.
With VirScan, scientists can run a single test to determine which viruses have infected an individual, rather than limiting their analysis to particular viruses. That unbiased approach could uncover unexpected factors affecting individual patients’ health, and also expands opportunities to analyze and compare viral infections in large populations.
The comprehensive analysis can be performed for about $25 per blood sample, but the test is currently being used only as a research tool and is not yet commercially available.
Stephen J. Elledge, an HHMI investigator at Brigham and Women’s Hospital, led the development of VirScan. He and his colleagues have already used VirScan to screen the blood of 569 people in the United States, South Africa, Thailand, and Peru. The scientists described the new technology and reported their findings in the June 5, 2015, issue of the journal Science.
Virus antibodies: clues that last decades
Bacteriophage (credit: Wikimedia Commons)
VirScan works by screening the blood for antibodies against any of the 206 species of viruses known to infect humans*. The immune system ramps up production of pathogen-specific antibodies when it encounters a virus for the first time, and it can continue to produce those antibodies for years or decades after it clears an infection.
That means VirScan not only identifies viral infections that the immune system is actively fighting, but also provides a history of an individual’s past infections.
To develop the new test, Elledge and his colleagues synthesized more than 93,000 short pieces of DNA encoding different segments of viral proteins. They introduced those pieces of DNA into bacteria-infecting viruses called bacteriophage.
Each bacteriophage manufactured one of the protein segments — known as a peptide — and displayed the peptide on its surface. As a group, the bacteriophage displayed all of the protein sequences found in the more than 1,000 known strains of human viruses.
To test the method, the team used it to analyze blood samples from patients known to be infected with particular viruses, including HIV and hepatitis C. “It turns out that it works really well,” Elledge says. “We were in the sensitivity range of 95 to 100 percent for those, and the specificity was good—we didn’t falsely identify people who were negative. That gave us confidence that we could detect other viruses, and when we did see them we would know they were real.”
Harvard Medical School | Viral History in a Drop of Blood
International study
Elledge and his colleagues used VirScan to analyze the antibodies in 569 people from four countries, examining about 100 million potential antibody/epitope interactions. They found that on average, each person had antibodies to ten different species of viruses. As expected, antibodies against certain viruses were common among adults but not in children, suggesting that children had not yet been exposed to those viruses. Individuals residing South Africa, Peru, and Thailand, tended to have antibodies against more viruses than people in the United States. The researchers also found that people infected with HIV had antibodies against many more viruses than did people without HIV.
Elledge says the team was surprised to find that antibody responses against specific viruses were surprisingly similar between individuals, with different people’s antibodies recognizing identical amino acids in the viral peptides. “In this paper alone we identified more antibody/peptide interactions to viral proteins than had been identified in the previous history of all viral exploration,” he says. The surprising reproducibility of those interactions allowed the team to refine their analysis and improve the sensitivity of VirScan, and Elledge says the method will continue to improve as his team analyzes more samples. Their findings on viral epitopes may also have important implications for vaccine design.
Elledge says the approach his team has developed is not limited to antiviral antibodies. His own lab is also using it to look for antibodies that attack a body’s own tissue in certain autoimmune diseases that are associated with cancer. A similar approach could also be used to screen for antibodies against other types of pathogens.
* Antibodies in the blood find their viral targets by recognizing unique features known as epitopes that are embedded in proteins on the virus surface. To perform the VirScan analysis, all of the peptide-displaying bacteriophage are allowed to mingle with a blood sample. Antiviral antibodies in the blood find and bind to their target epitopes within the displayed peptides. The scientists then retrieve the antibodies and wash away everything except for the few bacteriophage that cling to them. By sequencing the DNA of those bacteriophage, they can identify which viral protein pieces were grabbed onto by antibodies in the blood sample. That tells the scientists which viruses a person’s immune system has previously encountered, either through infection or through vaccination. Elledge estimates it would take about 2-3 days to process 100 samples, assuming sequencing is working optimally. He is optimistic the speed of the assay will increase with further development.
Abstract of Comprehensive serological profiling of human populations using a synthetic human virome
The human virome plays important roles in health and immunity. However, current methods for detecting viral infections and antiviral responses have limited throughput and coverage. Here, we present VirScan, a high-throughput method to comprehensively analyze antiviral antibodies using immunoprecipitation and massively parallel DNA sequencing of a bacteriophage library displaying proteome-wide peptides from all human viruses. We assayed over 108 antibody-peptide interactions in 569 humans across four continents, nearly doubling the number of previously established viral epitopes. We detected antibodies to an average of 10 viral species per person and 84 species in at least two individuals. Although rates of specific virus exposure were heterogeneous across populations, antibody responses targeted strongly conserved “public epitopes” for each virus, suggesting that they may elicit highly similar antibodies. VirScan is a powerful approach for studying interactions between the virome and the immune system.