
Eggnog should be your go-to holiday tipple. Here's how to make it safe, and delicious.
The post How to Use Science to Make Safe Eggnog With Raw Eggs appeared first on WIRED.

Science and reality

Eggnog should be your go-to holiday tipple. Here's how to make it safe, and delicious.
The post How to Use Science to Make Safe Eggnog With Raw Eggs appeared first on WIRED.
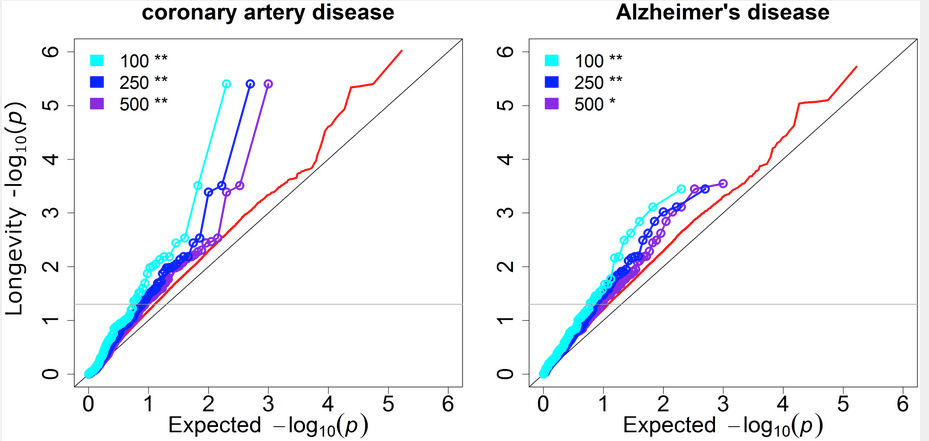
Disease GWAS show substantial genetic overlap with longevity. Shown are results for coronary artery disease and Alzheimer’s disease. The y axis is the observed P values for longevity, and the x axis is the expected P values under the null hypothesis that the disease is independent of longevity. The cyan, blue and purple lines show the P values for longevity of the top 100, 250, and 500 disease SNPs from independent genetic loci, respectively. Red lines show the background distribution of longevity P values for all independent genetic loci tested in both the longevity and disease GWAS. The grey horizontal line corresponds to the threshold for nominal significance (P< = 0.05) for longevity. Significance of enrichment was determined with the hypergeometric test. (credit: Kristen Fortney et al./PLOS Genetics)
What’s the secret of centenarians who have health and diet habits similar to the average person but have remained active and alert at very old ages?
Genes. That’s according to scientists at Stanford University and the University of Bologna, who have written a new report published in PLOS Genetics, based on their finding of several disease variants that may be absent in centenarians compared to the general population.
Genetic studies so far have only identified a single gene (APOE, known to be involved in Alzheimer’s disease) that is different in centenarians versus normal agers.
Finding additional longevity genes
To find the additional longevity genes, the authors first developed a new statistical method called informed GWAS (genome-wide association studies), which uses knowledge from 14 diseases to narrow down the search genes associated with longevity.
Using iGWAS, the scientists found eight SNPs (single nucleotide polymorphisms — molecular variations at different locations on the gene) that are significant for the centenarians they studied, and they were able to validate four of these in replication studies of long-lived subjects.
The four “longevity loci” (gene locations) along with the APOE gene may provide clues about physiological mechanisms for successful aging. These loci are known to be involved in various processes including cell senescence, autoimmunity, and cell signaling, and also with Alzheimer’s disease.
The incidence of nearly all diseases increases with age, so understanding genetic factors for successful aging could have a large impact on health. Future work may lead to a better understanding of how these genes promote successful aging and could identify additional longevity genes by recruiting more centenarians for analysis.
We developed a new statistical framework to find genetic variants associated with extreme longevity. The method, informed GWAS (iGWAS), takes advantage of knowledge from large studies of age-related disease in order to narrow the search for SNPs associated with longevity. To gain support for our approach, we first show there is an overlap between loci involved in disease and loci associated with extreme longevity. These results indicate that several disease variants may be depleted in centenarians versus the general population. Next, we used iGWAS to harness information from 14 meta-analyses of disease and trait GWAS to identify longevity loci in two studies of long-lived humans. In a standard GWAS analysis, only one locus in these studies is significant (APOE/TOMM40) when controlling the false discovery rate (FDR) at 10%. With iGWAS, we identify eight genetic loci to associate significantly with exceptional human longevity at FDR < 10%. We followed up the eight lead SNPs in independent cohorts, and found replication evidence of four loci and suggestive evidence for one more with exceptional longevity. The loci that replicated (FDR < 5%) includedAPOE/TOMM40 (associated with Alzheimer’s disease), CDKN2B/ANRIL (implicated in the regulation of cellular senescence), ABO (tags the O blood group), and SH2B3/ATXN2 (a signaling gene that extends lifespan in Drosophila and a gene involved in neurological disease). Our results implicate new loci in longevity and reveal a genetic overlap between longevity and age-related diseases and traits, including coronary artery disease and Alzheimer’s disease. iGWAS provides a new analytical strategy for uncovering SNPs that influence extreme longevity, and can be applied more broadly to boost power in other studies of complex phenotypes.
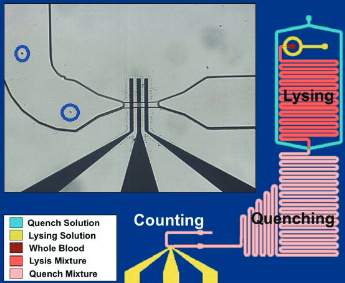
Schematic of the leukocyte counting chip with lysing, quenching, and counter modules shown in different colors. The insert (upper left) is an enlarged view of the platinum microfabricated electrodes (yellow). (credit: U. Hassan et al./TECHNOLOGY)
A microfluidic biosensor that can count red blood cells, platelets, and white blood cells electrically using just one drop of blood (11 microL) has been developed by University of Illinois at Urbana-Champaign (UIUC) researchers, replacing the standard hematology analyzer, a large, expensive lab device that requires trained technicians and physical sample transportation.
The new biosensor can electrically count the different types of blood cells based on their size and membrane properties. To count leukocyte and its differentials, red blood cells are selectively lysed and the remaining white blood cells were individually counted. Specific cells like neutrophils are counted using multi-frequency analysis, which probe the membrane properties of the cells.
The device, which will use credit-card-size disposable cartridges, requires minimal or no experience. It is expected to find uses in hospitals at the bedside, private clinics, retail clinics, and the developing world.
Patients can perform the test at home in under 20 minutes and share the results with their primary care physicians electronically, reducing the cost of the test to less than $10, compared to $100 or more currently, says UIUc Professor Rashid Bashir, principal investigator.
The research appears in the December 2015 issue of the journal TECHNOLOGY
Abstract of A microfluidic biochip for complete blood cell counts at the point-of-care
Complete blood cell counts (CBCs) are one of the most commonly ordered and informative blood tests in hospitals. The results from a CBC, which typically include white blood cell (WBC) counts with differentials, red blood cell (RBC) counts, platelet counts and hemoglobin measurements, can have implications for the diagnosis and screening of hundreds of diseases and treatments. Bulky and expensive hematology analyzers are currently used as a gold standard for acquiring CBCs. For nearly all CBCs performed today, the patient must travel to either a hospital with a large laboratory or to a centralized lab testing facility. There is a tremendous need for an automated, portable point-of-care blood cell counter that could yield results in a matter of minutes from a drop of blood without any trained professionals to operate the instrument. We have developed microfluidic biochips capable of a partial CBC using only a drop of whole blood. Total leukocyte and their 3-part differential count are obtained from 10 μL of blood after on-chip lysing of the RBCs and counting of the leukocytes electrically using microfabricated platinum electrodes. For RBCs and platelets, 1 μL of whole blood is diluted with PBS on-chip and the cells are counted electrically. The total time for measurement is under 20 minutes. We demonstrate a high correlation of blood cell counts compared to results acquired with a commercial hematology analyzer. This technology could potentially have tremendous applications in hospitals at the bedside, private clinics, retail clinics and the developing world.
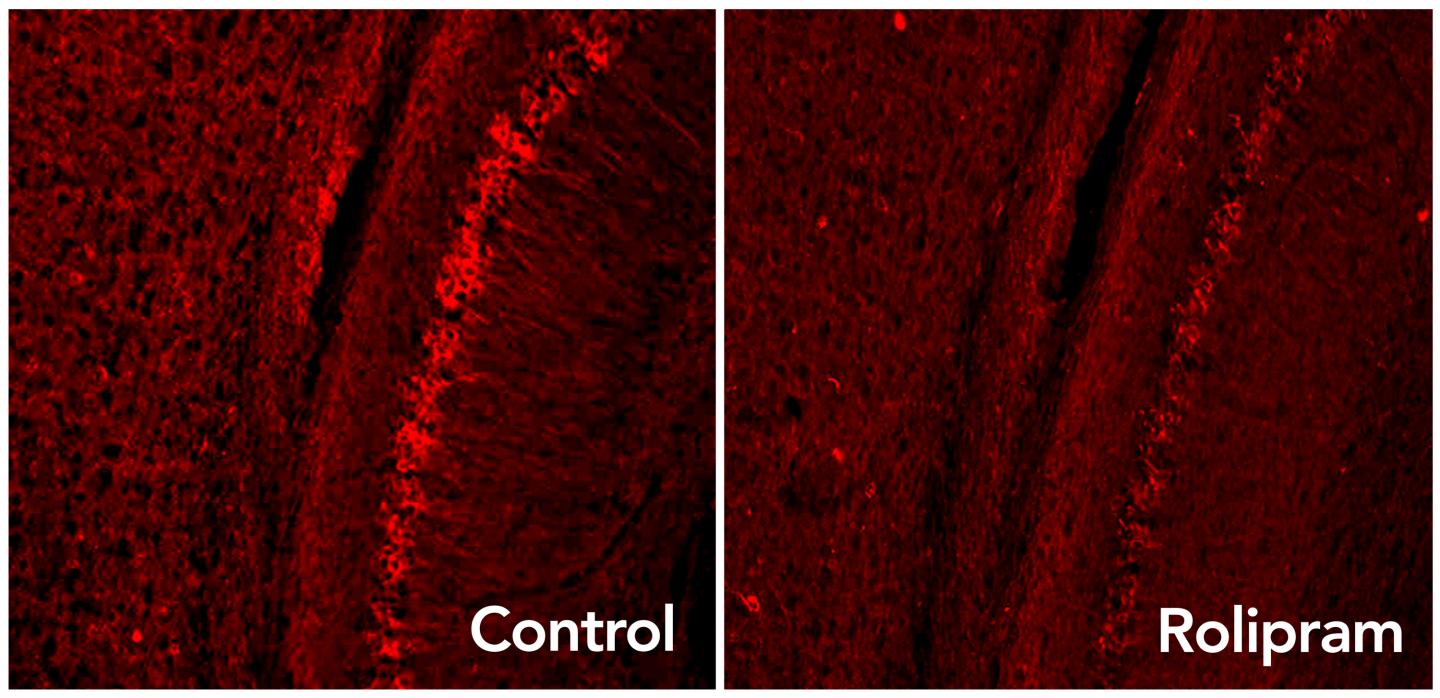
Rolipram drug activates the brain’s garbage disposal system, eliminating excess tau proteins (glowing red dots) associated with neurodegenerative diseases such as Alzheimer’s. (credit: Laboratory of Karen Duff/Columbia University Medical Center)
Rolipram, a drug that boosts activity in the brain’s “garbage disposal” system, can decrease levels of toxic proteins associated with Alzheimer’s disease and other neurodegenerative disorders and improve cognition in mice, a new study by neuroscientists has found.
Rolipram causes nausea, but similar drugs do not, and could be tested in clinical trials quickly, the researchers say.
“This has the potential to open up new avenues of treatment for Alzheimer’s and many other neurodegenerative diseases,” said study leader Karen E. Duff, PhD, professor of pathology and cell biology at Columbia University Medical Center (CUMC) and New York State Psychiatric Institute (NYSPI).
A “garbage-disposal” switch
To remain healthy, brain cells must continually clear out old, worn, or damaged proteins. This task is performed by a small molecule called the proteasome, which works like a kitchen garbage-disposal system, grinding up the old proteins so they can be recycled into new ones. However, in neurodegenerative diseases such as Alzheimer’s, Parkinson’s, and Huntington’s, proteins tagged for destruction accumulate in the brain’s neurons. This suggests that the cell’s proteasomes are impaired.
The cause for this: tau — a protein that accumulates in Alzheimer’s and other brain diseases — sticks to the proteasome and jams up the protein garbage-disposal process, the researchers first discovered (using a genetically engineered mouse).
In the new research, administering rolipram activated the proteasome and restored protein disposal. The drug also improved memory in diseased mice to levels seen in healthy mice.
Rolipram has been tested before in mice, and was shown to improve memory. But the new research shows a previously unknown function of the drug: it produces a physical change in the proteasome and increases its activity.
Should ‘clear out everything at once’
Duff says we still don’t know exactly which form of a particular protein is toxic to the brain, which has made it difficult to develop drugs to treat neurodegenerative diseases. “In Alzheimer’s disease, the problem is compounded because several types of abnormal protein can accumulate in a person’s brain, including amyloid, tau, alpha-synuclein, and TDP43.
However, the researchers think that “a well-functioning proteasome will be able to clear out everything at once,” she says — including Alzheimer’s, frontotemporal degeneration, Huntington’s, and Parkinson’s.
The study was published Tuesday (Dec. 22) in the online edition of Nature Medicine. The National Institute of Health’s National Institute of Neurological Disorders and Stroke provided funding for the study.
The ubiquitin proteasome system (UPS) degrades misfolded proteins including those implicated in neurodegenerative diseases. We investigated the effects of tau accumulation on proteasome function in a mouse model of tauopathy and in a cross to a UPS reporter mouse (line Ub-G76V-GFP). Accumulation of insoluble tau was associated with a decrease in the peptidase activity of brain 26S proteasomes, higher levels of ubiquitinated proteins and undegraded Ub-G76V-GFP. 26S proteasomes from mice with tauopathy were physically associated with tau and were less active in hydrolyzing ubiquitinated proteins, small peptides and ATP. 26S proteasomes from normal mice incubated with recombinant oligomers or fibrils also showed lower hydrolyzing capacity in the same assays, implicating tau as a proteotoxin. Administration of an agent that activates cAMP–protein kinase A (PKA) signaling led to attenuation of proteasome dysfunction, probably through proteasome subunit phosphorylation. In vivo, this led to lower levels of aggregated tau and improvements in cognitive performance.

This year's planetary research reminds us that exploration of the solar system (and beyond) is just beginning.
The post All the Year’s Kickbutt Science From Space appeared first on WIRED.

Looking at the top stories from the WIRED science team this year, you might get the feeling that we're a little down on the future of planet Earth.
The post Drought, Anti-Vaxxers, and Cancer Made For a Great 2015 appeared first on WIRED.
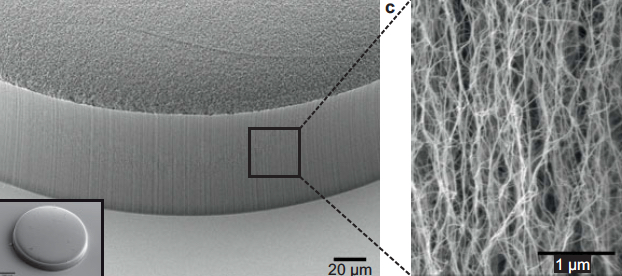
Scanning electron microscope image of carbon nanotubes showing textured porosity (credit: Allison L. Yost et al./Microsystems & Nanoengineering)
Engineers at MIT have devised a new technique for trapping hard-to-detect molecules, using forests of coated carbon nanotubes.
The team modified a simple microfluidic channel with an array of vertically aligned carbon nanotubes — rolled lattices of carbon atoms that resemble tiny tubes of chicken wire.
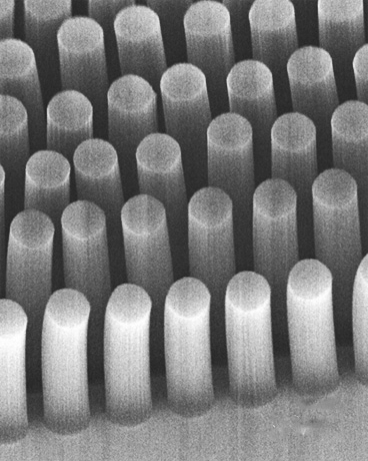
Carbon-nanotube posts can trap cancer and other cells as they flow through a microfluidic device (credit: Brian Wardle)
The researchers had previously devised a method for standing carbon nanotubes on their ends, like trees in a forest (see “Trapping cancer cells with carbon nanotubes“). This 3-D array of permeable carbon nanotubes allows fluid to flow through a microfluidic device.
Now, in a study published this week in the Journal of Microengineering and Nanotechnology, the researchers have given the nanotube array the additional ability to trap specific particles. To do this, the team coated the array, layer by layer, with polymers of alternating electric charge.
Depending on the number of layers deposited, the researchers can create thicker or thinner nanotubes and thereby tailor the porosity of the forest to capture larger or smaller particles of interest. The nanotubes’ polymer coating can also be chemically manipulated to bind specific bioparticles flowing through the forest.
The combination of carbon nanotubes and multilayer coatings may help finely tune microfluidic devices to capture extremely small and rare particles, such as certain viruses and proteins, says Brian Wardle, professor of aeronautics and astronautics at MIT.
“There are smaller bioparticles that contain very rich amounts of information that we don’t currently have the ability to access in point-of-care [medical testing] devices like microfluidic chips,” says Wardle, who is a co-author on the paper. “Carbon nanotube arrays could actually be a platform that could target that size of bioparticle.”
What’s more, Wardle says, a three-dimensional forest of carbon nanotubes would provide much more surface area on which target molecules may interact, compared with the two-dimensional surfaces in conventional microfluidics.
Capturing specific particles of interest
To test this idea, the researchers used an established technique to treat the surface of the nanotubes with antibodies that bind to prostate specific antigen (PSA), a common experimental target.
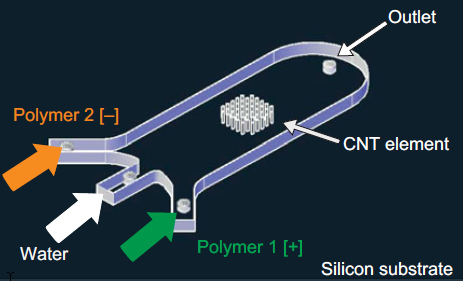
A 3-D array of carbon nanotubes on a microfluidic device coated with successive layers of alternately charged polymer solutions (credit: Allison L. Yost et al./Microsystems & Nanoengineering)
The team integrated a 3-D array of carbon nanotubes into a microfluidic device by using chemical vapor deposition and photolithography to grow and pattern carbon nanotubes onto silicon wafers. They then grouped the nanotubes into a cylinder-shaped forest, measuring about 50 micrometers tall and 1 millimeter wide, and centered the array within a 3 millimeter-wide, 7-millimeter long microfluidic channel.
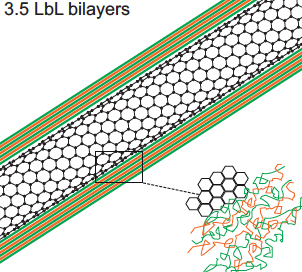
Polyelectrolyte multilayer (PEM) film deposition on carbon-nanotube surface (credit: Allison L. Yost et al./Microsystems & Nanoengineering)
The researchers coated the nanotubes in successive layers of alternately charged polymer solutions to create distinct, binding layers around each nanotube. To do so, they flowed each solution through the channel and found they were able to create a more uniform coating with a gap between the top of the nanotube forest and the roof of the channel. Such a gap allowed solutions to flow over, then down into the forest, coating each individual nanotube.
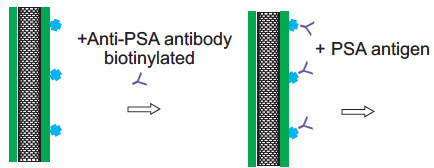
Carbon nanotube treated with antibodies for PSA capture (credit: Allison L. Yost et al./Microsystems & Nanoengineering)
After coating the nanotube array in layers of polymer solution, the researchers demonstrated that the array could be primed to detect a given molecule by treating it with antibodies that typically bind to prostate specific antigen (PSA). They pumped in a solution containing small amounts of PSA and found that the array captured the antigen effectively, throughout the forest, rather than just on the outer surface of a typical microfluidic element.
The polymer-coated arrays captured 40 percent more antigens, compared with arrays lacking the polymer coating.
Wardle says that the nanotube array is extremely versatile. The carbon nanotubes can be manipulated mechanically, electrically, and optically and the polymer coatings can be chemically altered to capture a wide range of particles. He says an immediate target may be biomarkers called exosomes, which are less than 100 nanometers wide and can be important signals of a disease’s progression.
“This type of device actually has all the characteristics and functionality that would allow you to go after bioparticles like exosomes and things that really truly are nanometer scale,” he noted.
This research was funded in part by the National Science Foundation.
We demonstrate the layer-by-layer (LbL) assembly of polyelectrolyte multilayers (PEM) on three-dimensional nanofiber scaffolds. High porosity (99%) aligned carbon nanotube (CNT) arrays are photolithographically patterned into elements that act as textured scaffolds for the creation of functionally coated (nano)porous materials. Nanometer-scale bilayers of poly(allylamine hydrochloride)/poly(styrene sulfonate) (PAH/SPS) are formed conformally on the individual nanotubes by repeated deposition from aqueous solution in microfluidic channels. Computational and experimental results show that the LbL deposition is dominated by the diffusive transport of the polymeric constituents, and we use this understanding to demonstrate spatial tailoring on the patterned nanoporous elements. A proof-of-principle application, microfluidic bioparticle capture using N-hydroxysuccinimide-biotin binding for the isolation of prostate-specific antigen (PSA), is demonstrated.
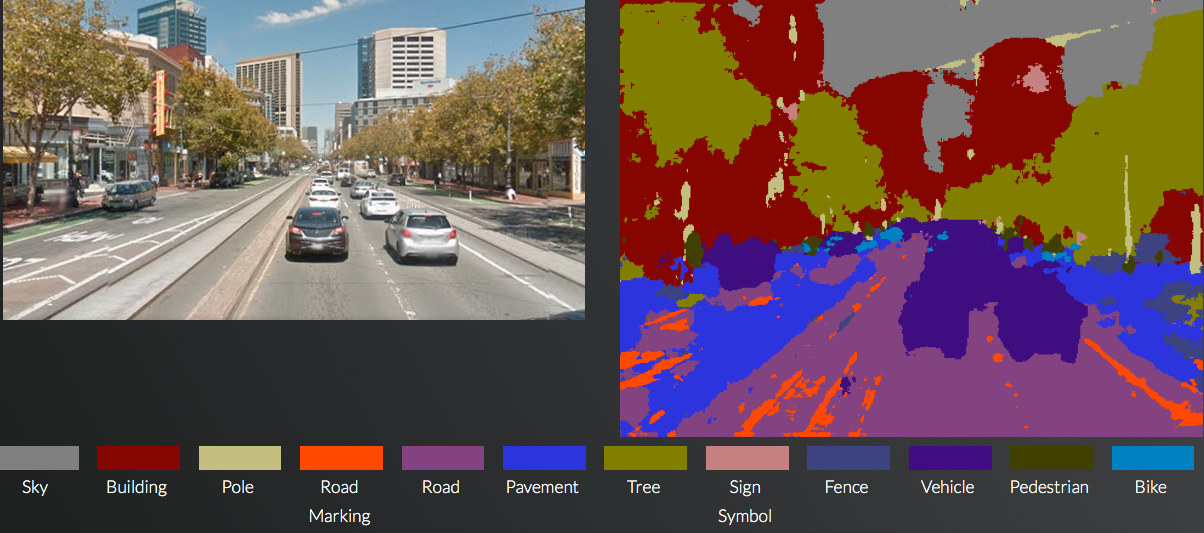
In a test by KurzweilAI using a Google Maps image of Market Street In San Francisco, the SegNet system accurately identified the various elements, even hard-to-see pedestrians (shown in brown on the left) and road markings. (credit: KurzweilAI/Cambridge University/Google)
Two new technologies that use deep-learning techniques to help machines see and analyze images (such as roads and people) could improve visual performance for driveless cars and create a new generation of smarter smartphones and cameras.
Designed by University of Cambridge researchers, the systems can recognize their own location and surroundings. Most driverless cars currently in development use radar and LIDAR sensors, which often cost more than the car itself. (See “New laser design could dramatically shrink autonomous-vehicle 3-D laser-ranging systems” for another solution.)
One of the systems, SegNet, can identify a user’s location and orientation, including places where GPS does not function, and can identify the various components of a road scene in real time on a regular camera or smartphone (see image above or try it yourself here).
SegNet can take an image of a street scene it hasn’t seen before and classify it, sorting objects into 12 different categories — such as roads, street signs, pedestrians, buildings and cyclists — in real time. It can deal with light, shadow, and night-time environments, and currently labels more than 90% of pixels correctly, according to the researchers. Previous systems using expensive laser or radar based sensors have not been able to reach this level of accuracy while operating in real time, the researchers say.
To create SegNet, Cambridge undergraduate students manually labeled every pixel in each of 5000 images, with each image taking about 30 minutes to complete. Once the labeling was finished, the researchers “trained” the system, which was successfully tested on both city roads and motorways.
“It’s remarkably good at recognizing things in an image, because it’s had so much practice,” said Alex Kendall, a PhD student in the Department of Engineering. “However, there are a million knobs that we can turn to fine-tune the system so that it keeps getting better.”
SegNet was primarily trained in highway and urban environments, so it still has some learning to do for rural, snowy, or desert environments. The system is not yet at the point where it can be used to control a car or truck, but it could be used as a warning system, similar to the anti-collision technologies currently available on some passenger cars.
But teaching a machine to see is far more difficult than it sounds, said Professor Roberto Cipolla, who led the research. “There are three key technological questions that must be answered to design autonomous vehicles: where am I, what’s around me and what do I do next?”
SegNet addresses the second question. The researchers’ Visual Localization system answers the first question. Using deep learning, it can determine their location and orientation from a single color image in a busy urban scene. The researchers say the system is far more accurate than GPS and works in places where GPS does not, such as indoors, in tunnels, or in cities where a reliable GPS signal is not available.
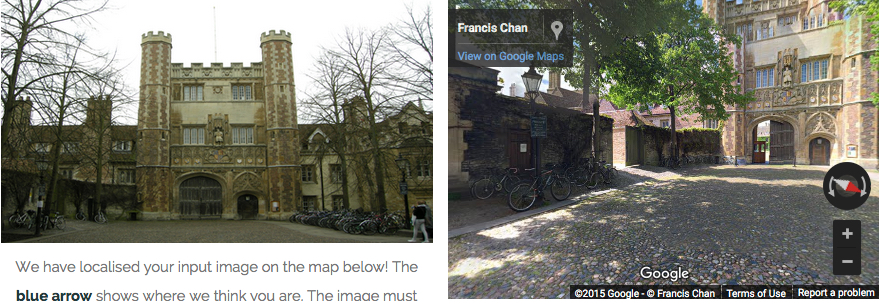
In a KurzweilAI test of the Visual Localization system (using an image in the Central Cambridge UK demo), the system accurately identified a Cambridge building, displaying the correct Google Maps street view, and marked its location on a Google map (credit: KurzweilAI/Cambridge University/Google)
It has been tested along a kilometer-long stretch of King’s Parade in central Cambridge, and it is able to determine both location and orientation within a few meters and a few degrees, which is far more accurate than GPS — a vital consideration for driverless cars, according to the researchers. (Try it here.)
The localization system uses the geometry of a scene to learn its precise location, and is able to determine, for example, whether it is looking at the east or west side of a building, even if the two sides appear identical.
“In the short term, we’re more likely to see this sort of system on a domestic robot — such as a robotic vacuum cleaner, for instance,” said Cipolla. “It will take time before drivers can fully trust an autonomous car, but the more effective and accurate we can make these technologies, the closer we are to the widespread adoption of driverless cars and other types of autonomous robotics.”
The researchers are presenting details of the two technologies at the International Conference on Computer Vision in Santiago, Chile.
Cambridge University | Teaching machines to see
We present a robust and real-time monocular six degree of freedom relocalization system. Our system trains a convolutional neural network to regress the 6-DOF camera pose from a single RGB image in an end-to-end manner with no need of additional engineering or graph optimisation. The algorithm can operate indoors and outdoors in real time, taking 5ms per frame to compute. It obtains approximately 2m and 3◦accuracy for large scale outdoor scenes and 0.5m and 5◦accuracy indoors. This is achieved using an efficient 23 layer deep convnet, demonstrating that convnets can be used to solve complicated out of image plane regression problems. This was made possible by leveraging transfer learning from large scale classi- fication data. We show that the PoseNet localizes from high level features and is robust to difficult lighting, motion blur and different camera intrinsics where point based SIFT registration fails. Furthermore we show how the pose feature that is produced generalizes to other scenes allowing us to regress pose with only a few dozen training examples.